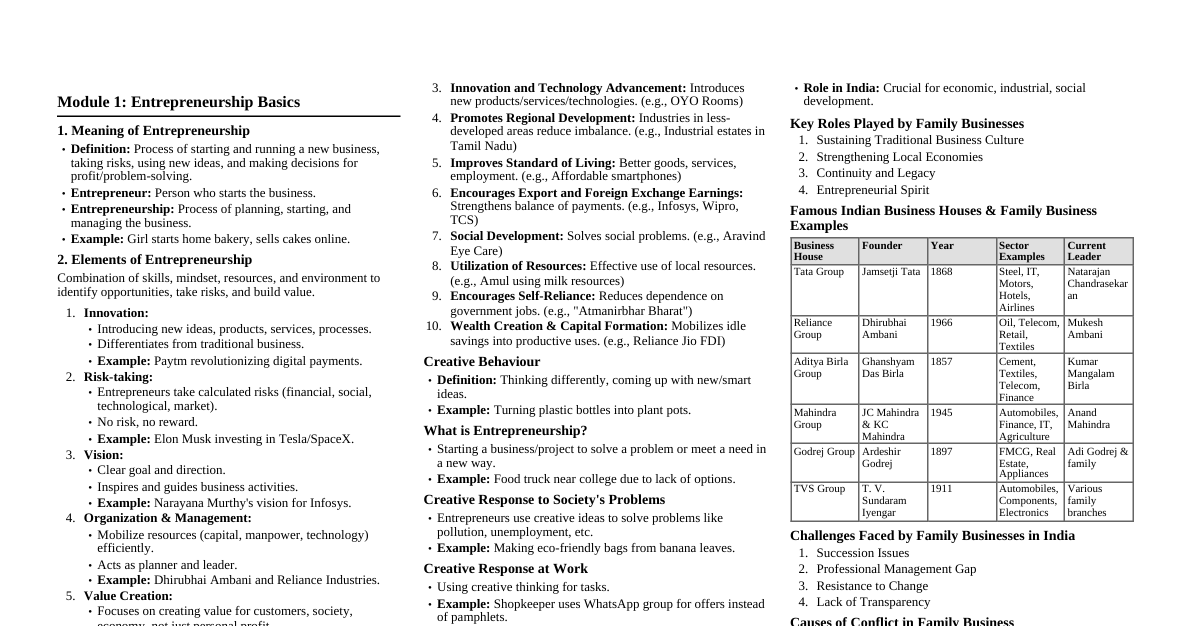
Polymerase Chain Reaction (PCR) Definition: A molecular biology technique used to amplify a single copy or a few copies of a piece of DNA across several orders of magnitude, generating thousands to millions of copies of a particular DNA sequence. Invented by: Kary Mullis in 1983. Key Components of a PCR Reaction DNA Template: The DNA sequence to be amplified. DNA Polymerase: An enzyme (e.g., Taq polymerase) that synthesizes new DNA strands. Must be thermostable. Primers: Short, single-stranded DNA sequences (typically 18-24 nucleotides) that are complementary to the ends of the target DNA region. Two primers are needed (forward and reverse). Deoxynucleotide Triphosphates (dNTPs): Building blocks (A, T, C, G) for the new DNA strands. Buffer Solution: Provides optimal conditions (pH, salt concentration) for the DNA polymerase activity. Often contains $Mg^{2+}$ ions, which are a co-factor for Taq polymerase. Thermal Cycler: An instrument that rapidly changes the temperature of the reaction mixture. PCR Cycle Steps Denaturation: Temperature: $94-98^\circ C$ (typically $95^\circ C$) for $15-30$ seconds. Purpose: Heating the reaction mixture separates the double-stranded DNA template into single strands by breaking hydrogen bonds. Annealing: Temperature: $50-65^\circ C$ (typically $55-60^\circ C$) for $15-60$ seconds. This temperature is specific to the primers (melting temperature, $T_m$). Purpose: Primers bind (anneal) to their complementary sequences on the single-stranded DNA template strands. Extension (Elongation): Temperature: $70-74^\circ C$ (typically $72^\circ C$) for $10$ seconds to several minutes, depending on the length of the target DNA. (Taq polymerase optimal activity). Purpose: DNA polymerase synthesizes new DNA strands by adding dNTPs complementary to the template, starting from the primers and extending in the $5' \rightarrow 3'$ direction. These three steps are repeated for $25-40$ cycles, leading to exponential amplification of the target DNA. PCR Reaction Profile Initial Denaturation: 95°C for 2-5 min (ensures all DNA is denatured) Cycling (25-40 cycles): Denaturation: 95°C for 15-30 sec Annealing: 50-65°C for 15-60 sec Extension: 72°C for 10 sec - several min Final Extension: 72°C for 5-10 min (ensures all remaining single-stranded DNA is extended) Final Hold: 4-10°C (short-term storage of products) Calculation of Target DNA Copies After $n$ cycles, the theoretical number of target DNA copies is $2^n$. Example: After $30$ cycles, $2^{30} \approx 1$ billion copies. Types of PCR Reverse Transcription PCR (RT-PCR): Amplifies DNA from an RNA template using reverse transcriptase to synthesize cDNA first. Quantitative PCR (qPCR) / Real-time PCR: Monitors the amplification of a target DNA molecule during the PCR reaction in real-time, allowing quantification of initial DNA template. Uses fluorescent dyes or probes. Nested PCR: Uses two sets of primers to amplify a single target sequence. The second set of primers binds inside the product of the first PCR to increase specificity and sensitivity. Multiplex PCR: Amplifies multiple target DNA sequences simultaneously in a single reaction using multiple sets of primers. Gradient PCR: Uses a thermal cycler with a temperature gradient across the block to test different annealing temperatures in a single run, optimizing reaction conditions. Hot Start PCR: Reduces non-specific amplification by preventing DNA polymerase activity during reaction setup at room temperature, often using antibodies or chemical modifications. Applications of PCR Gene Cloning: Amplifying specific genes for insertion into vectors. DNA Sequencing: Preparing DNA templates for sequencing. Genetic Fingerprinting: Forensic analysis, paternity testing. Disease Diagnosis: Detecting pathogens (viruses, bacteria), genetic disorders. Mutation Detection: Identifying specific mutations in genes. Gene Expression Analysis: (via RT-qPCR) Quantifying RNA levels. Evolutionary Studies: Amplifying ancient DNA for phylogenetic analysis. Factors Affecting PCR Success Primer Design: Specificity, $T_m$, lack of secondary structures (hairpins, primer-dimers). DNA Polymerase Activity: Enzyme concentration, buffer composition ($Mg^{2+}$). Template Quality/Quantity: Purity, concentration, presence of inhibitors. Thermal Cycler Accuracy: Precise temperature control and ramp rates. Contamination: Presence of foreign DNA can lead to false positives. Primer Design Considerations Length: $18-24$ base pairs (bp). GC Content: $40-60\%$ (affects $T_m$). Melting Temperature ($T_m$): Primers in a pair should have similar $T_m$ (within $5^\circ C$). Annealing temperature is typically $2-5^\circ C$ below $T_m$. Avoid Secondary Structures: Hairpins, self-dimers, cross-dimers. Specificity: Should bind only to the target sequence. 3' End Stability: Avoid runs of more than three G or C bases at the 3' end to prevent non-specific priming.