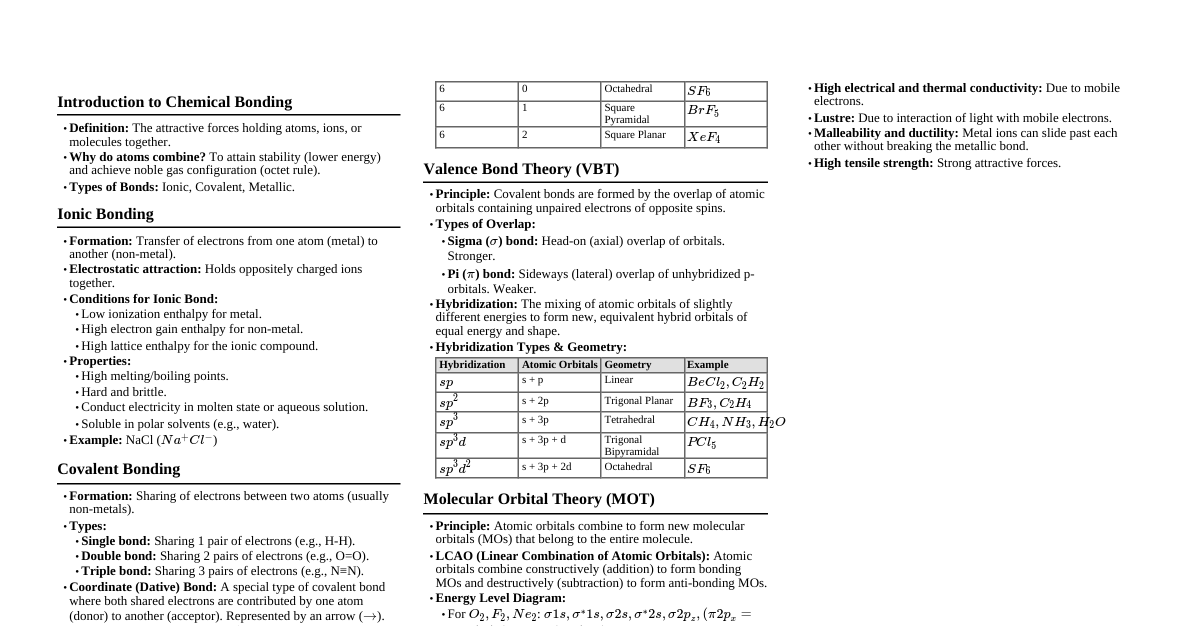
1. Types of Chemical Bonds Ionic Bond: Formed by complete transfer of electrons. Between metals (low IE) and non-metals (high EN, high EA). Forms ions: Cation (+) and Anion (-). Lattice energy is key for stability. Covalent Bond: Formed by mutual sharing of electrons. Between two non-metals. Can be single, double, or triple bonds. Polarity depends on electronegativity difference ($\Delta EN$). $\Delta EN = 0$: Non-polar. $0 $\Delta EN \ge 1.7$: Ionic (approximate value). Coordinate Bond (Dative Bond): A special type of covalent bond where both shared electrons come from one atom (donor). Requires a donor (with lone pair) and an acceptor (with empty orbital). Example: $\text{NH}_4^+$, $\text{H}_3\text{O}^+$, $\text{CO}$. Metallic Bond: Between metal atoms in a metallic solid. "Sea of electrons" model: Delocalized electrons shared among metal cations. Responsible for metallic properties (conductivity, malleability, ductility). 2. Lewis Structures & Formal Charge Lewis Dot Structure: Represents valence electrons as dots. Octet Rule: Atoms tend to gain, lose, or share electrons to achieve 8 valence electrons (except H, Li, Be, B). Exceptions: Incomplete octet (e.g., $\text{BF}_3$), Expanded octet (e.g., $\text{SF}_6$, $\text{PCl}_5$), Odd-electron molecules (e.g., $\text{NO}$). Formal Charge (FC): $$ \text{FC} = (\text{Valence electrons}) - (\text{Non-bonding electrons}) - \frac{1}{2}(\text{Bonding electrons}) $$ The sum of FC in a molecule/ion equals its total charge. Prefer structures with FC as close to zero as possible. Negative FC on more electronegative atoms is preferred. 3. Valence Shell Electron Pair Repulsion (VSEPR) Theory Predicts molecular geometry based on minimizing electron pair repulsion. Electron domains: Lone pairs (LP) and bond pairs (BP). Multiple bonds (double/triple) count as one electron domain. Order of repulsion: $\text{LP-LP} > \text{LP-BP} > \text{BP-BP}$. Steric No. LP BP Electron Geometry Molecular Geometry Example 2 0 2 Linear Linear $\text{BeCl}_2$, $\text{CO}_2$ 3 0 3 Trigonal Planar Trigonal Planar $\text{BF}_3$, $\text{SO}_3$ 3 1 2 Trigonal Planar Bent $\text{SO}_2$, $\text{O}_3$ 4 0 4 Tetrahedral Tetrahedral $\text{CH}_4$, $\text{CCl}_4$ 4 1 3 Tetrahedral Trigonal Pyramidal $\text{NH}_3$, $\text{PCl}_3$ 4 2 2 Tetrahedral Bent $\text{H}_2\text{O}$, $\text{SF}_2$ 5 0 5 Trigonal Bipyramidal Trigonal Bipyramidal $\text{PCl}_5$ 5 1 4 Trigonal Bipyramidal Seesaw $\text{SF}_4$ 5 2 3 Trigonal Bipyramidal T-shaped $\text{ClF}_3$ 5 3 2 Trigonal Bipyramidal Linear $\text{XeF}_2$ 6 0 6 Octahedral Octahedral $\text{SF}_6$ 6 1 5 Octahedral Square Pyramidal $\text{BrF}_5$ 6 2 4 Octahedral Square Planar $\text{XeF}_4$ 4. Hybridization Mixing of atomic orbitals to form new hybrid orbitals with equivalent energy and shape. Steric Number (SN) = (Number of sigma bonds) + (Number of lone pairs). SN Hybridization Geometry 2 $sp$ Linear 3 $sp^2$ Trigonal Planar 4 $sp^3$ Tetrahedral 5 $sp^3d$ Trigonal Bipyramidal 6 $sp^3d^2$ Octahedral Sigma ($\sigma$) bonds: Formed by head-on overlap. All single bonds are $\sigma$. Pi ($\pi$) bonds: Formed by sideways overlap of unhybridized p-orbitals. Present in double (one $\pi$) and triple (two $\pi$) bonds. 5. Molecular Orbital Theory (MOT) Atomic orbitals combine to form molecular orbitals (MOs). Bonding MOs (lower energy, constructive interference) and Antibonding MOs (higher energy, destructive interference). Filling order for diatomic molecules ($Z \le 7$): $\sigma1s, \sigma^*1s, \sigma2s, \sigma^*2s, \pi2p_x = \pi2p_y, \sigma2p_z, \pi^*2p_x = \pi^*2p_y, \sigma^*2p_z$ Filling order for diatomic molecules ($Z > 7$): $\sigma1s, \sigma^*1s, \sigma2s, \sigma^*2s, \sigma2p_z, \pi2p_x = \pi2p_y, \pi^*2p_x = \pi^*2p_y, \sigma^*2p_z$ Bond Order (BO): $$ \text{BO} = \frac{1}{2} (\text{Number of electrons in bonding MOs} - \text{Number of electrons in antibonding MOs}) $$ BO $> 0$: Stable molecule. Higher BO: Stronger bond, shorter bond length. Paramagnetic: Unpaired electrons in MOs. Diamagnetic: All electrons paired in MOs. 6. Dipole Moment ($\mu$) Measure of polarity of a molecule. $$ \mu = q \times d $$ where $q$ is charge magnitude and $d$ is distance. Vector quantity. Net dipole moment determines overall molecular polarity. Symmetrical molecules often have $\mu = 0$ (non-polar) even if individual bonds are polar (e.g., $\text{CO}_2$, $\text{CCl}_4$, $\text{BF}_3$). Asymmetrical molecules usually have $\mu \ne 0$ (polar) (e.g., $\text{H}_2\text{O}$, $\text{NH}_3$, $\text{CHCl}_3$). 7. Hydrogen Bonding Special type of dipole-dipole interaction. Occurs when H is bonded to a highly electronegative atom (F, O, N). Involves a H atom between two electronegative atoms, one covalently bonded to H (donor), the other attracting H (acceptor). Types: Intermolecular H-bonding: Between different molecules (e.g., $\text{H}_2\text{O}$, $\text{HF}$, alcohols). Increases boiling point, viscosity. Intramolecular H-bonding: Within the same molecule (e.g., o-nitrophenol, salicylaldehyde). Decreases boiling point, increases volatility. 8. Resonance When a single Lewis structure cannot adequately describe the bonding in a molecule or ion. Involves delocalization of $\pi$ electrons or lone pairs. Actual structure is a resonance hybrid of contributing canonical forms. Resonance hybrid is more stable than any single canonical form (resonance energy). Example: $\text{O}_3$, $\text{CO}_3^{2-}$, $\text{NO}_3^-$, Benzene. Resonance Rules: Only position of electrons (lone pairs, $\pi$ bonds) can change, not atoms. Number of paired/unpaired electrons must remain constant. All contributing structures must be valid Lewis structures. Contributing structures should have similar energy. 9. Bond Parameters Bond Length: Average distance between nuclei of two bonded atoms. Factors: Size of atoms (larger atoms, longer bond), Bond multiplicity (triple Bond Energy/Enthalpy: Energy required to break one mole of a specific bond. Higher bond order, higher bond energy. Correlates inversely with bond length. Bond Angle: Angle between orbitals containing bonding electron pairs around central atom. Determined by molecular geometry (VSEPR and hybridization). Lone pair repulsion causes deviation from ideal angles.